I work on the integration of multi-omics data, using and implementing state of the art analysis methods for the understanding of gene regulation and therapeutic target discovery. I am currently working as a postdoc at the Biomedical Genomics Group @ Health Data Science Unit at the BioQuant Center and Medical Faculty Heidelberg.
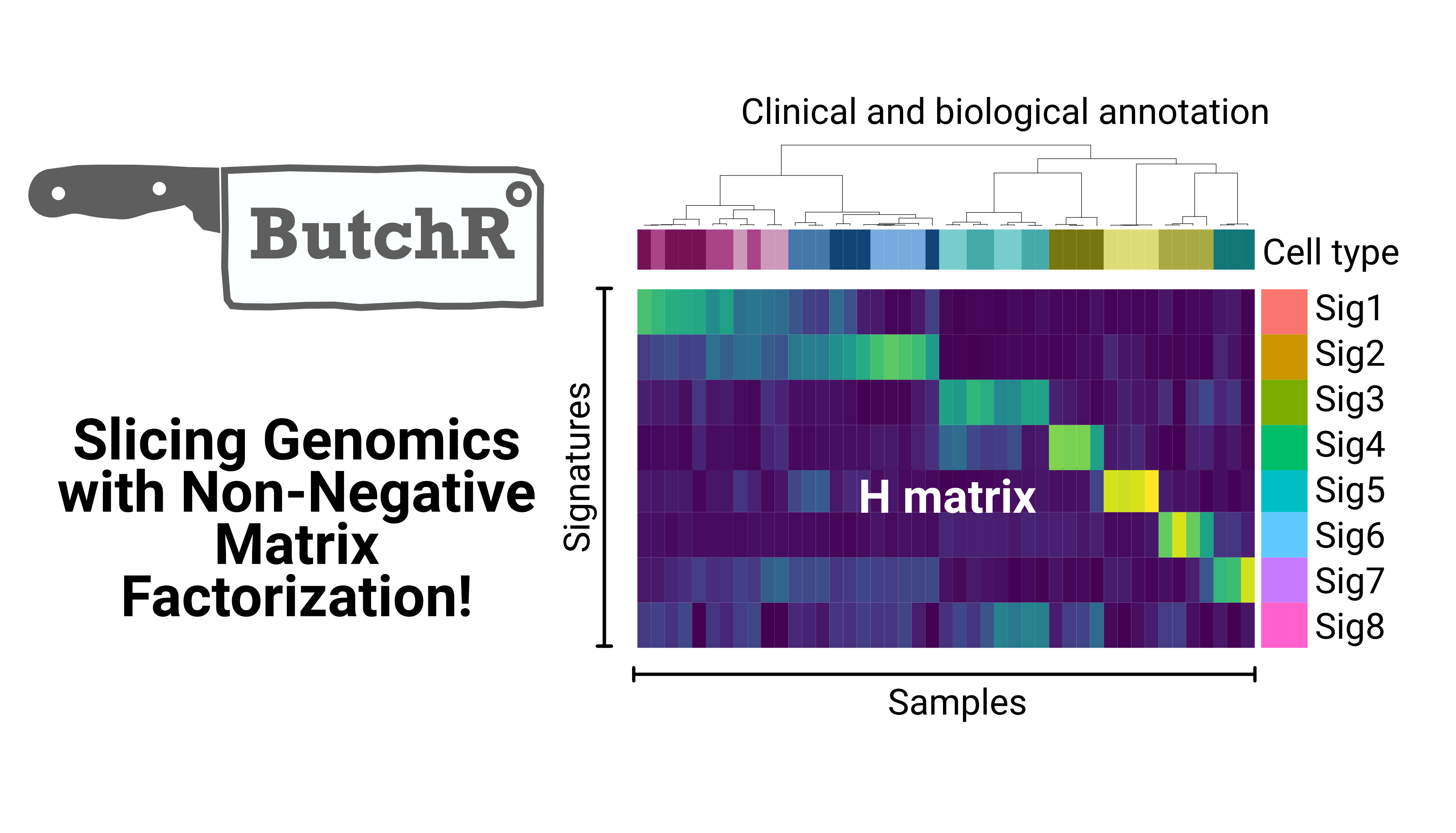
During my doctoral studies at the Health Data Science Unit, I developed ButchR, a new toolkit to infer signatures and extract relevant features associated with genotypes and phenotypes using non-negative matrix factorization (NMF). This tool can be used to extract a lower dimensional representation from multi-omics data layers in bulk and single-cell genomics.
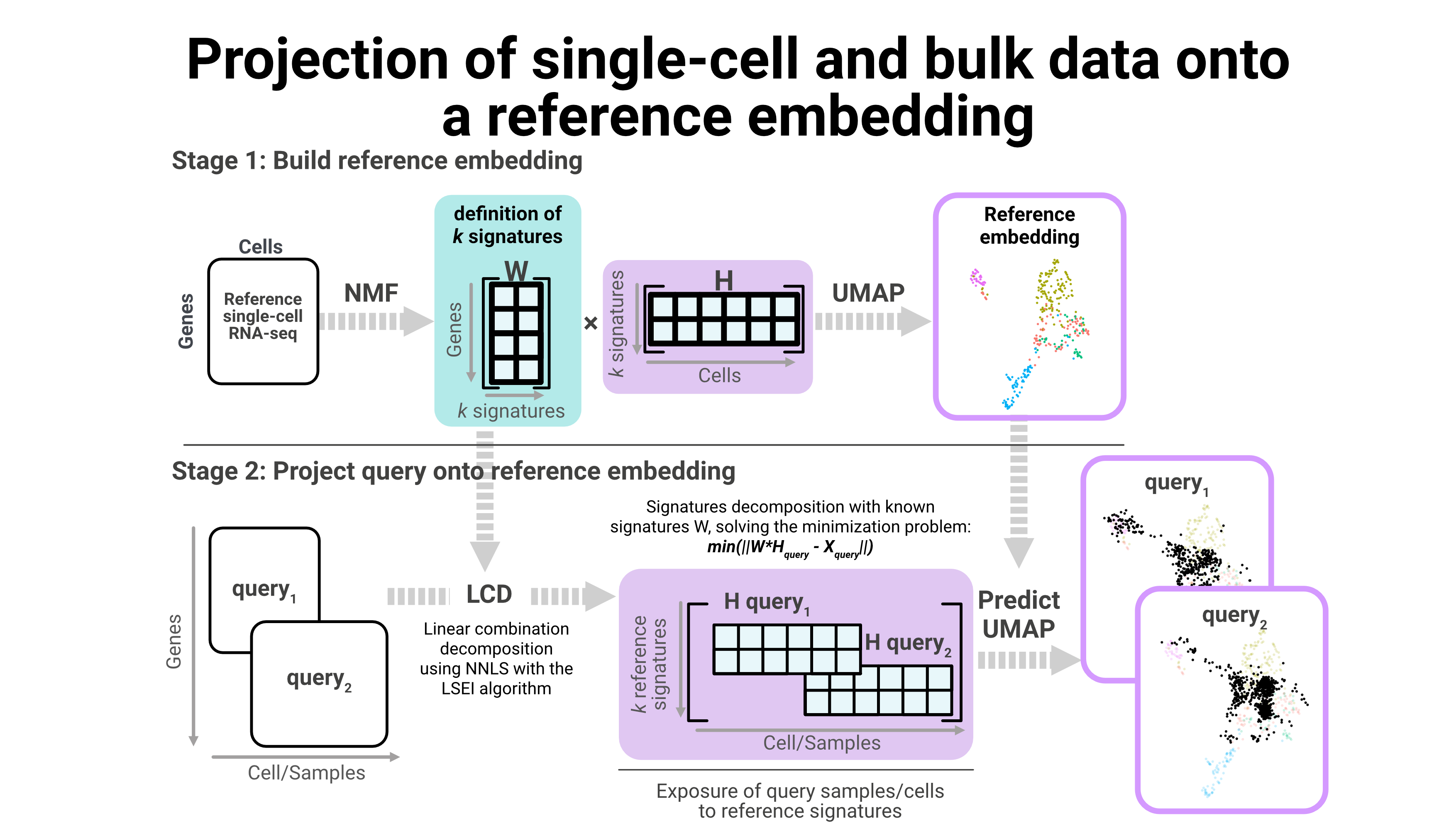 I am also developing new strategies to combine single-cell and bulk data, like projecting samples and cells into a reference embedding. Using this approach we have been able to understand the possible cell of origin for complex tumor identities.
I am also developing new strategies to combine single-cell and bulk data, like projecting samples and cells into a reference embedding. Using this approach we have been able to understand the possible cell of origin for complex tumor identities.
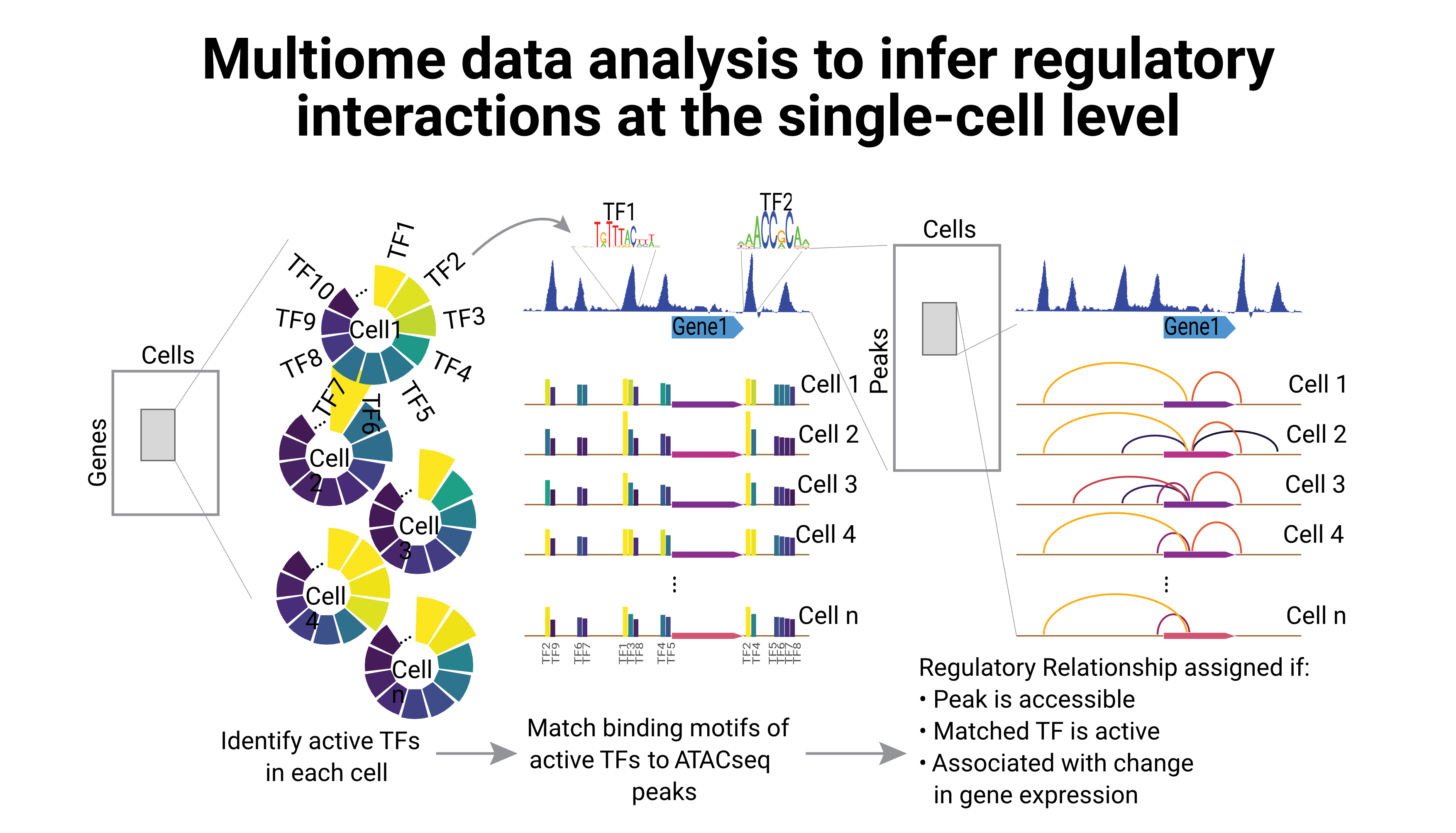 Regulation of gene expression is tightly coupled with the epigenetic landscape of the cell; but until recently, it was not possible to study the direct relationship between the transcriptome and the chromatin accessibility at the single-cell level. However, several multiome technologies have overcome this limitation, providing both layers of information within the same single cell.
Regulation of gene expression is tightly coupled with the epigenetic landscape of the cell; but until recently, it was not possible to study the direct relationship between the transcriptome and the chromatin accessibility at the single-cell level. However, several multiome technologies have overcome this limitation, providing both layers of information within the same single cell.
One of my main research interests is to use such data to infer the regulatory interactions between one gene and its cis-regulatory elements. I am currently using single-cell multiomics data to infer the regulatory landscape on single-cells, in order to identify regulatory differences between different cell types.
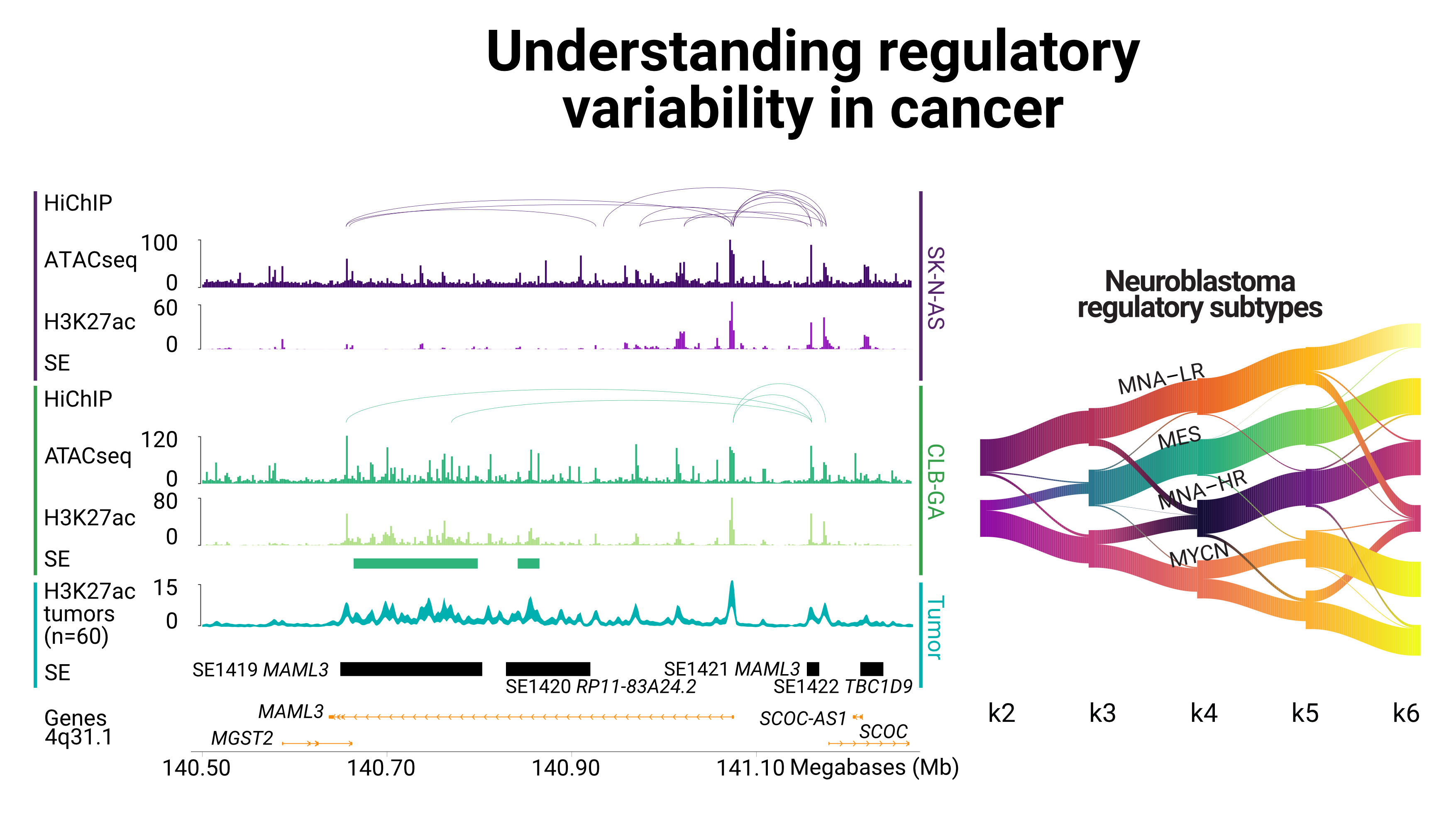 Neuroblastoma subtypes appear to be strongly defined through epigenomic profiles. In close collaboration with the Division of Neuroblastoma Genomics at the DKFZ and the HDSU team at bioquant, I worked in the integration of ChIP-seq data on histone modification to extract specific signatures for NB subtypes from tumors and cell lines.
Neuroblastoma subtypes appear to be strongly defined through epigenomic profiles. In close collaboration with the Division of Neuroblastoma Genomics at the DKFZ and the HDSU team at bioquant, I worked in the integration of ChIP-seq data on histone modification to extract specific signatures for NB subtypes from tumors and cell lines.