(For a full list of publications see below)
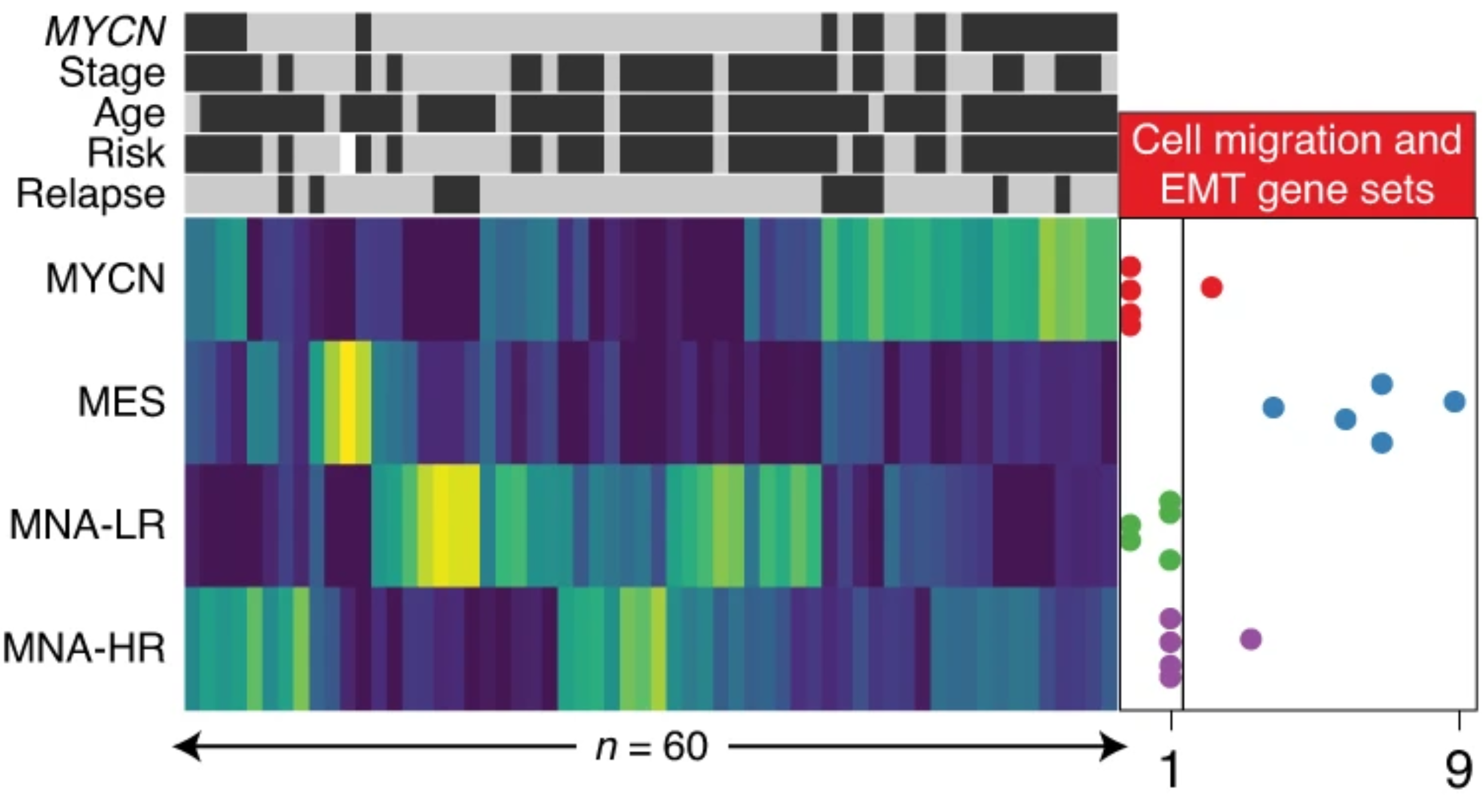
Using genome-wide H3K27ac profiles across 60 NBs, we identified four major super-enhancer-driven epigenetic subtypes and their underlying master regulatory networks. Three of these subtypes recapitulated known clinical groups, while the fourth subtype, exhibiting mesenchymal characteristics, ressemblance to Schwann cell precursors, was induced by RAS activation and was enriched in relapsed disease. This study reveals subtype-specific super-enhancer regulation in NBs.
Moritz Gartlgruber*, Ashwini Kumar Sharma*, Andrés Quintero*, Daniel Dreidax*, Selina Jansky, Young-Gyu Park, Sina Kreth, Johanna Meder, Daria Doncevic, Paul Saary, Umut H Toprak, Naveed Ishaque, Elena Afanasyeva, Elisa Wecht, Jan Koster, Rogier Versteeg, Thomas GP Grünewald, David TW Jones, Stefan M Pfister, Kai-Oliver Henrich, Johan van Nes, Carl Herrmann*, Frank Westermann*

Non-negative matrix factorization (NMF) has been widely used for the analysis of genomic data to perform feature extraction and signature identification due to the interpretability of the decomposed signatures. However, running a basic NMF analysis requires the installation of multiple tools and dependencies, along with a steep learning curve and computing time. To mitigate such obstacles, we developed ShinyButchR, a novel R/Shiny application that provides a complete NMF-based analysis workflow, allowing the user to perform matrix decomposition using NMF, feature extraction, interactive visualization, relevant signature identification, and association to biological and clinical variables. ShinyButchR builds upon the also novel R package ButchR, which provides new TensorFlow solvers for algorithms of the NMF family, functions for downstream analysis, a rational method to determine the optimal factorization rank and a novel feature selection strategy.
Andres Quintero, Daniel Hübschmann, Nils Kurzawa, Sebastian Steinhauser, Philipp Rentzsch, Stephen Krämer, Carolin Andresen, Jeongbin Park, Roland Eils, Matthias Schlesner, Carl Herrmann
Biology Methods and Protocols (2020)
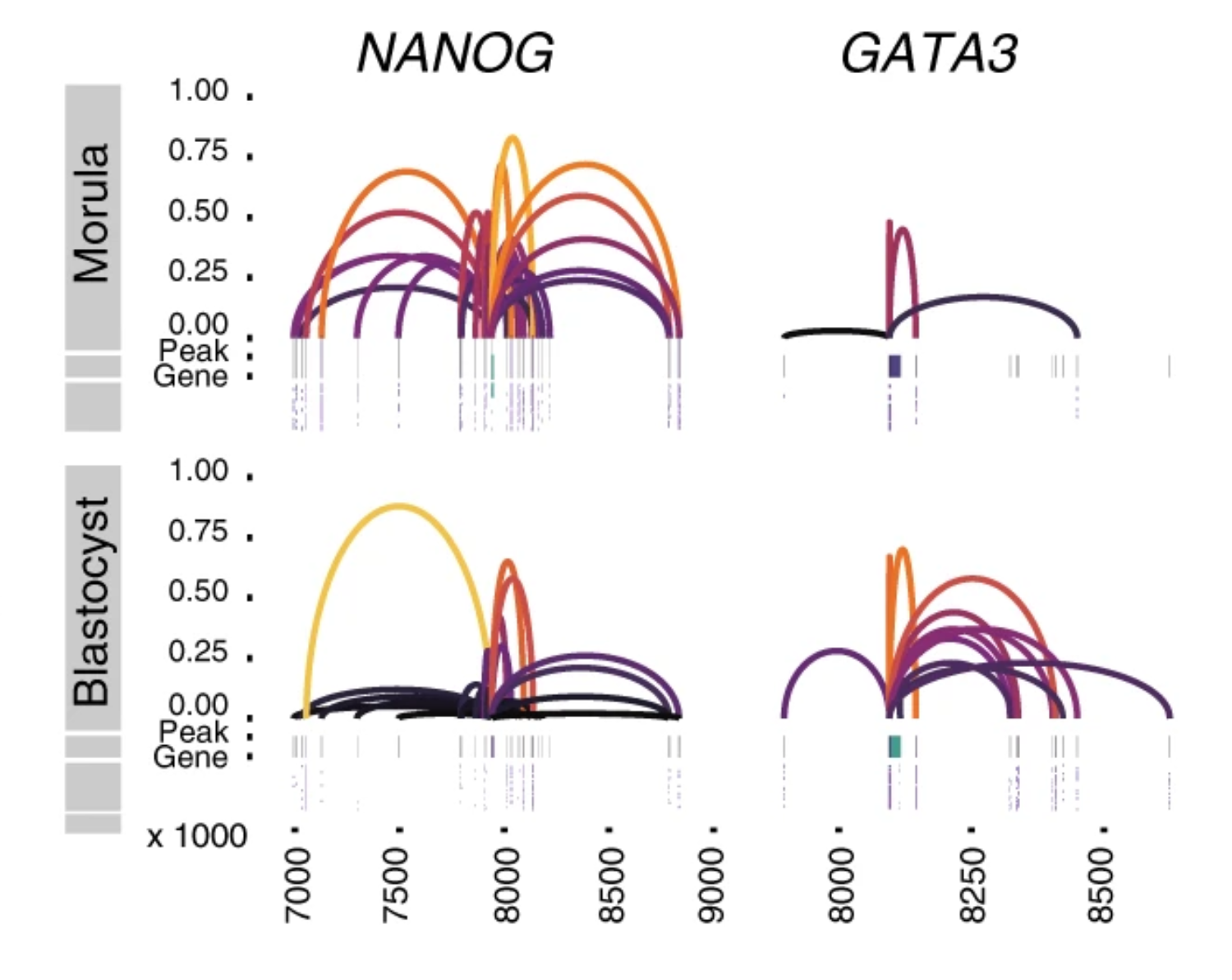
Integrative analysis of multi-omics layers at single cell level is critical for accurate dissection of cell-to-cell variation within certain cell populations. Here we report scCAT-seq, a technique for simultaneously assaying chromatin accessibility and the transcriptome within the same single cell. We show that the combined single cell signatures enable accurate construction of regulatory relationships between cis-regulatory elements and the target genes at single-cell resolution, providing a new dimension of features that helps direct discovery of regulatory patterns specific to distinct cell identities. Moreover, we generate the first single cell integrated map of chromatin accessibility and transcriptome in early embryos and demonstrate the robustness of scCAT-seq in the precise dissection of master transcription factors in cells of distinct states. The ability to obtain these two layers of omics data will help provide more accurate definitions of “single cell state” and enable the deconvolution of regulatory heterogeneity from complex cell populations.
Longqi Liu, Chuanyu Liu, Andrés Quintero, Liang Wu, Yue Yuan, Mingyue Wang, Mengnan Cheng, Lizhi Leng, Liqin Xu, Guoyi Dong, Rui Li, Yang Liu, Xiaoyu Wei, Jiangshan Xu, Xiaowei Chen, Haorong Lu, Dongsheng Chen, Quanlei Wang, Qing Zhou, Xinxin Lin, Guibo Li, Shiping Liu, Qi Wang, Hongru Wang, J Lynn Fink, Zhengliang Gao, Xin Liu, Yong Hou, Shida Zhu, Huanming Yang, Yunming Ye, Ge Lin, Fang Chen, Carl Herrmann, Roland Eils, Zhouchun Shang, Xun Xu
Single-cell transcriptomic analyses provide insights into the developmental origins of neuroblastoma
Selina Jansky, Ashwini Kumar Sharma, Verena Körber, Andrés Quintero, Umut H Toprak, Elisa M Wecht, Moritz Gartlgruber, Alessandro Greco, Elad Chomsky, Thomas GP Grünewald, Kai-Oliver Henrich, Amos Tanay, Carl Herrmann, Thomas Höfer, Frank Westermann
Nature genetics (2021)
Identifying multimodal signatures underlying the somatic comorbidity of psychosis: the COMMITMENT roadmap
Emanuel Schwarz, Dag Alnæs, Ole A Andreassen, Han Cao, Junfang Chen, Franziska Degenhardt, Daria Doncevic, Dominic Dwyer, Roland Eils, Jeanette Erdmann, Carl Herrmann, Martin Hofmann-Apitius, Tobias Kaufmann, Nikolaos Koutsouleris, Alpha T Kodamullil, Adyasha Khuntia, Sören Mucha, Markus M Nöthen, Riya Paul, Mads L Pedersen, Andres Quintero, Heribert Schunkert, Ashwini Sharma, Heike Tost, Lars T Westlye, Youcheng Zhang, Andreas Meyer-Lindenberg
Molecular Psychiatry (2021)
Super enhancers define regulatory subtypes and cell identity in neuroblastoma
Moritz Gartlgruber*, Ashwini Kumar Sharma*, Andrés Quintero*, Daniel Dreidax*, Selina Jansky, Young-Gyu Park, Sina Kreth, Johanna Meder, Daria Doncevic, Paul Saary, Umut H Toprak, Naveed Ishaque, Elena Afanasyeva, Elisa Wecht, Jan Koster, Rogier Versteeg, Thomas GP Grünewald, David TW Jones, Stefan M Pfister, Kai-Oliver Henrich, Johan van Nes, Carl Herrmann*, Frank Westermann*
Nature Cancer (2021)
ShinyButchR: Interactive NMF-based decomposition workflow of genome-scale datasets
Andres Quintero, Daniel Hübschmann, Nils Kurzawa, Sebastian Steinhauser, Philipp Rentzsch, Stephen Krämer, Carolin Andresen, Jeongbin Park, Roland Eils, Matthias Schlesner, Carl Herrmann
Biology Methods and Protocols (2020)
Deconvolution of single-cell multi-omics layers reveals regulatory heterogeneity
Longqi Liu, Chuanyu Liu, Andrés Quintero, Liang Wu, Yue Yuan, Mingyue Wang, Mengnan Cheng, Lizhi Leng, Liqin Xu, Guoyi Dong, Rui Li, Yang Liu, Xiaoyu Wei, Jiangshan Xu, Xiaowei Chen, Haorong Lu, Dongsheng Chen, Quanlei Wang, Qing Zhou, Xinxin Lin, Guibo Li, Shiping Liu, Qi Wang, Hongru Wang, J Lynn Fink, Zhengliang Gao, Xin Liu, Yong Hou, Shida Zhu, Huanming Yang, Yunming Ye, Ge Lin, Fang Chen, Carl Herrmann, Roland Eils, Zhouchun Shang, Xun Xu
Nature communications (2019)